In this post, I attempt to call a Rust function from an R session and compare the runtime of that function to a similar function written in R.
The rextendr R library allows us to call rust functions from an R session. As the repository linked above mentions, you “need to setup a working Rust toolchain”.
I write a Rust function to get the \(n^{th}\) Fibonacci term, and then compile it using the rust_source() function from the rextendr library, as shown below:
library(rextendr)
library(parallel)
# write code to calculate the nth fibonacci term
rust_code <- "
#[extendr]
fn fibonacci_rust(n:i32) -> i64 {
let mut fibn:i64 = 0;
if n==0 {
return 0;
} else if n==1 {
return 1;
} else {
return fibonacci_rust(n-1) + fibonacci_rust(n-2);
}
}
"
# compile source code
rust_source(code = rust_code)
# run the rust function
rust_fn_result = fibonacci_rust(15)
# print the value
print(rust_fn_result)
## [1] 610
Which is the correct value of the \(15^{th}\) Fibonacci number. Note that the Rust code begins with the #[extendr] preamble, otherwise the function fibonacci_rust() will not be available in the R environment.
I now create a similar function in R:
# write r function
fibonacci <- function(n) {
if(n==0) {
return(0)
} else if(n==1) {
return(1)
} else {
return(fibonacci(n-1) + fibonacci(n-2))
}
}
# run the R function
r_fn_result = fibonacci(15)
# print the value
print(r_fn_result)
## [1] 610
Which is the correct value again. I now test the runtime of each function for a series of values starting from 1 and ending at 40. In addition, I also give Rust a handicap: I’ll run the R function in parallel, while Rust will run serially.
# set input values
n <- 1:40
# create functions that collect elapsed times
elapsed_time_rust <- function(x) {
system.time(fibonacci_rust(x))['elapsed']
}
elapsed_time_r <- function(x) {
system.time(fibonacci(x))['elapsed']
}
# call rust function, also note the time it takes to run for all inputs
rust_total_runtime <- system.time({
rust_times <- sapply(n, FUN = elapsed_time_rust)
})
# running the R function in parallel, since it will take longer as n gets larger
num_cores <- max(1, detectCores() - 1)
# Create a cluster with the specified number of worker processes
cl <- makeCluster(num_cores)
# Export the helper function to all workers
clusterExport(cl, varlist = c("fibonacci"))
# Use parLapply to apply the function in parallel
r_total_runtime <- system.time({
r_times <- parLapply(cl, n, fun = elapsed_time_r)
})
# Simplify the result into a vector
r_times <- unlist(r_times)
# stop cluster
stopCluster(cl)
# print times taken
print("Time taken to calculate all fibonacci terms by Rust (serially):")
print(rust_total_runtime)
print("Time taken to calculate all fibonacci terms by R (in parallel):")
print(r_total_runtime)
## [1] "Time taken to calculate all fibonacci terms by Rust (serially):"
## user system elapsed
## 6.028 0.008 6.035
## [1] "Time taken to calculate all fibonacci terms by R (in parallel):"
## user system elapsed
## 0.353 0.302 250.130
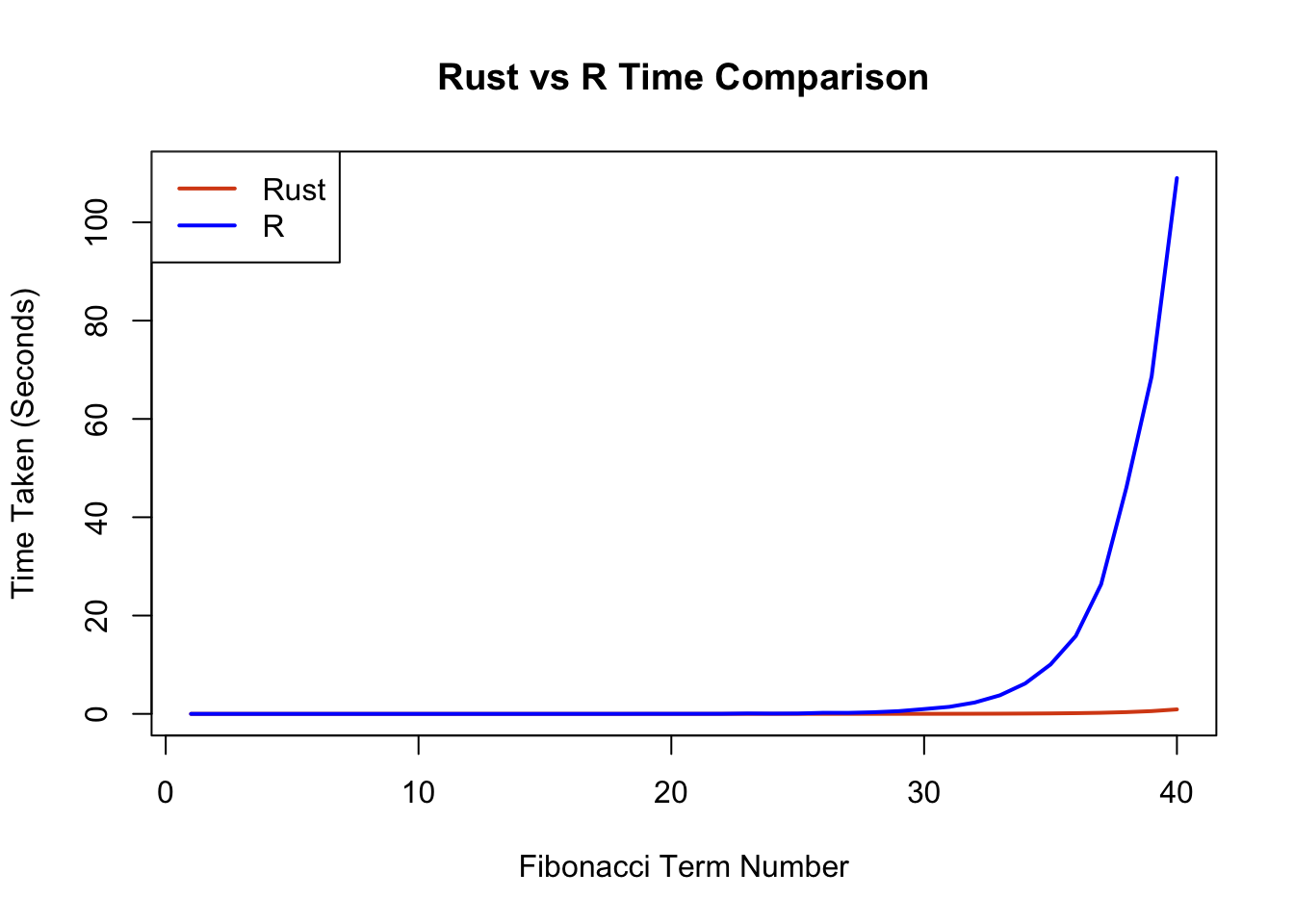
Rust takes 6 seconds to get the first 40 fibonacci terms in serial mode, while R takes 250 seconds (roughly 4 minutes) to do the same thing, in parallel! (Note: I used 11 cores on my machine for parallel execution).
Let’s also plot the time taken to calculate each term:
plot(n, rust_times, type = "l", col = "#D84B16", lwd = 2, xlab = "Fibonacci Term Number",
ylab = "Time Taken (Seconds)", main = "Rust vs R Time Comparison",
ylim = c(0, max(rust_times, r_times) + 1))
lines(n, r_times, col = "blue", lwd = 2)
# Add a legend with custom labels for each line
legend("topleft", legend = c("Rust", "R"), col = c("#D84B16", "blue"), lwd = 2)
While the example of the function I used is trivial, I can see other cases where it might be advantageous to migrate functions to Rust and then call those functions in R (better yet, just do everything in Rust if possible). I could also try optimizing the R function via memoization and then comparing its performance against its Rust counterpart. I do that in this post.